Fig. 2
From: A tale of two sequences: microRNA-target chimeric reads
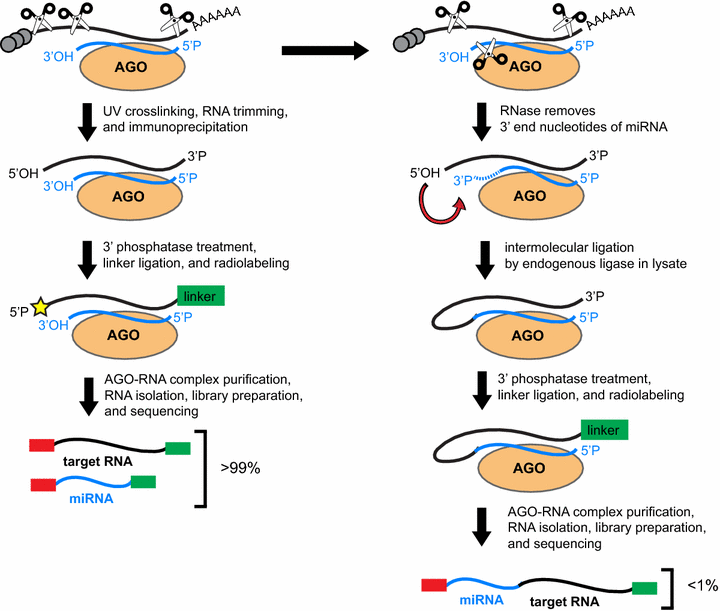
Comparison of the biochemical steps in CLIP-seq for the generation of standard CLIP-seq reads to events that can lead to the formation of miRNA-target chimeric reads in CLIP-seq or iPAR-CLIP. Standard CLIP-seq reads are generated after RNA trimming of UV crosslinked lysates and immunoprecipitation of the AGO-miRNA-target RNA tertiary complex. The 3′ end of the RNA is then prepared for linker ligation and the complex is radio-labeled to facilitate the isolation of the complex. Chimeric reads may form in CLIP-seq when partial digestion of the 3′ end of the miRNA by RNase during the RNA trimming step of CLIP-seq or iPAR-CLIP produces a 2′–3′ cyclic phosphate or a 3′ phosphate. Endogenous ligases in the lysate have been predicted to be responsible for ligation of the 3′ end of the digested miRNA to the 5′ phosphate of the target RNA. The subsequent steps that occur in the CLIP-seq protocol prepare the miRNA-target chimera for sequencing
